How to Read Wikipedia Data to Pandas Dataframe
Intro to data structures¶
We'll offset with a quick, non-comprehensive overview of the fundamental data structures in pandas to get you started. The central behavior about information types, indexing, and axis labeling / alignment apply across all of the objects. To become started, import NumPy and load pandas into your namespace:
In [i]: import numpy every bit np In [two]: import pandas equally pd Here is a basic tenet to keep in listen: data alignment is intrinsic. The link betwixt labels and information volition not be broken unless done so explicitly by you.
We'll give a brief intro to the data structures, then consider all of the broad categories of functionality and methods in split sections.
Series¶
Series is a 1-dimensional labeled assortment capable of holding any data blazon (integers, strings, floating point numbers, Python objects, etc.). The axis labels are collectively referred to every bit the index. The bones method to create a Serial is to call:
>>> s = pd . Series ( data , index = index ) Here, data can be many different things:
-
a Python dict
-
an ndarray
-
a scalar value (like 5)
The passed alphabetize is a list of centrality labels. Thus, this separates into a few cases depending on what information is:
From ndarray
If data is an ndarray, index must be the same length as data. If no index is passed, one will exist created having values [0, ..., len(data) - 1] .
In [3]: s = pd . Series ( np . random . randn ( 5 ), index = [ "a" , "b" , "c" , "d" , "e" ]) In [4]: south Out[4]: a 0.469112 b -0.282863 c -ane.509059 d -ane.135632 e i.212112 dtype: float64 In [5]: s . index Out[5]: Alphabetize(['a', 'b', 'c', 'd', 'e'], dtype='object') In [6]: pd . Series ( np . random . randn ( five )) Out[half-dozen]: 0 -0.173215 1 0.119209 2 -1.044236 3 -0.861849 4 -2.104569 dtype: float64 Note
pandas supports not-unique alphabetize values. If an functioning that does not support duplicate index values is attempted, an exception will be raised at that time. The reason for existence lazy is nearly all performance-based (there are many instances in computations, similar parts of GroupBy, where the index is not used).
From dict
Series tin can be instantiated from dicts:
In [7]: d = { "b" : 1 , "a" : 0 , "c" : 2 } In [viii]: pd . Series ( d ) Out[viii]: b 1 a 0 c 2 dtype: int64 Note
When the data is a dict, and an alphabetize is not passed, the Series alphabetize will be ordered by the dict's insertion gild, if y'all're using Python version >= iii.six and pandas version >= 0.23.
If you're using Python < 3.6 or pandas < 0.23, and an index is not passed, the Series index will be the lexically ordered listing of dict keys.
In the instance higher up, if you were on a Python version lower than iii.six or a pandas version lower than 0.23, the Series would be ordered by the lexical gild of the dict keys (i.e. ['a', 'b', 'c'] rather than ['b', 'a', 'c'] ).
If an index is passed, the values in data corresponding to the labels in the alphabetize volition be pulled out.
In [9]: d = { "a" : 0.0 , "b" : 1.0 , "c" : 2.0 } In [10]: pd . Series ( d ) Out[10]: a 0.0 b 1.0 c ii.0 dtype: float64 In [11]: pd . Series ( d , index = [ "b" , "c" , "d" , "a" ]) Out[xi]: b 1.0 c two.0 d NaN a 0.0 dtype: float64 Annotation
NaN (not a number) is the standard missing data marking used in pandas.
From scalar value
If data is a scalar value, an index must be provided. The value volition be repeated to match the length of index.
In [12]: pd . Serial ( 5.0 , index = [ "a" , "b" , "c" , "d" , "due east" ]) Out[12]: a 5.0 b v.0 c five.0 d v.0 e v.0 dtype: float64 Series is ndarray-like¶
Series acts very similarly to a ndarray , and is a valid argument to most NumPy functions. Notwithstanding, operations such as slicing will also piece the alphabetize.
In [13]: south [ 0 ] Out[13]: 0.4691122999071863 In [xiv]: southward [: 3 ] Out[14]: a 0.469112 b -0.282863 c -1.509059 dtype: float64 In [15]: south [ due south > southward . median ()] Out[15]: a 0.469112 e 1.212112 dtype: float64 In [16]: due south [[ iv , 3 , 1 ]] Out[16]: eastward ane.212112 d -ane.135632 b -0.282863 dtype: float64 In [17]: np . exp ( s ) Out[17]: a 1.598575 b 0.753623 c 0.221118 d 0.321219 east 3.360575 dtype: float64 Like a NumPy array, a pandas Series has a dtype .
In [18]: s . dtype Out[xviii]: dtype('float64') This is often a NumPy dtype. Nevertheless, pandas and 3rd-political party libraries extend NumPy's type system in a few places, in which case the dtype would be an ExtensionDtype . Some examples inside pandas are Categorical data and Nullable integer data type. Meet dtypes for more.
If yous need the actual array backing a Serial , use Series.array .
In [19]: s . array Out[xix]: <PandasArray> [ 0.4691122999071863, -0.2828633443286633, -1.5090585031735124, -1.1356323710171934, 1.2121120250208506] Length: v, dtype: float64 Accessing the array tin be useful when you need to do some operation without the alphabetize (to disable automatic alignment, for example).
Series.array volition ever be an ExtensionArray . Briefly, an ExtensionArray is a thin wrapper around i or more than physical arrays like a numpy.ndarray . pandas knows how to take an ExtensionArray and shop it in a Serial or a column of a DataFrame . See dtypes for more.
While Series is ndarray-like, if you lot need an actual ndarray, then utilize Series.to_numpy() .
In [20]: due south . to_numpy () Out[20]: array([ 0.4691, -0.2829, -1.5091, -i.1356, 1.2121]) Even if the Series is backed by a ExtensionArray , Series.to_numpy() will return a NumPy ndarray.
Series is dict-like¶
A Serial is like a fixed-size dict in that y'all can become and prepare values by alphabetize label:
In [21]: s [ "a" ] Out[21]: 0.4691122999071863 In [22]: s [ "east" ] = 12.0 In [23]: s Out[23]: a 0.469112 b -0.282863 c -one.509059 d -1.135632 e 12.000000 dtype: float64 In [24]: "east" in due south Out[24]: True In [25]: "f" in s Out[25]: False If a label is not independent, an exception is raised:
Using the get method, a missing label will return None or specified default:
In [26]: south . get ( "f" ) In [27]: due south . become ( "f" , np . nan ) Out[27]: nan See besides the section on aspect access.
Vectorized operations and label alignment with Serial¶
When working with raw NumPy arrays, looping through value-by-value is unremarkably not necessary. The aforementioned is true when working with Serial in pandas. Series can likewise be passed into virtually NumPy methods expecting an ndarray.
In [28]: s + s Out[28]: a 0.938225 b -0.565727 c -three.018117 d -ii.271265 e 24.000000 dtype: float64 In [29]: s * 2 Out[29]: a 0.938225 b -0.565727 c -3.018117 d -two.271265 e 24.000000 dtype: float64 In [thirty]: np . exp ( southward ) Out[30]: a 1.598575 b 0.753623 c 0.221118 d 0.321219 eastward 162754.791419 dtype: float64 A key difference between Series and ndarray is that operations between Series automatically align the information based on label. Thus, you can write computations without giving consideration to whether the Series involved have the same labels.
In [31]: south [ 1 :] + s [: - 1 ] Out[31]: a NaN b -0.565727 c -3.018117 d -2.271265 e NaN dtype: float64 The result of an operation betwixt unaligned Series will have the union of the indexes involved. If a label is non found in one Series or the other, the result will be marked as missing NaN . Being able to write code without doing whatsoever explicit data alignment grants immense freedom and flexibility in interactive data assay and research. The integrated data alignment features of the pandas information structures set pandas apart from the bulk of related tools for working with labeled data.
Note
In general, nosotros chose to brand the default result of operations between differently indexed objects yield the union of the indexes in order to avoid loss of information. Having an index label, though the data is missing, is typically of import information equally function of a ciphering. You lot of grade accept the selection of dropping labels with missing data via the dropna function.
Name attribute¶
Serial can also have a proper noun attribute:
In [32]: s = pd . Series ( np . random . randn ( five ), name = "something" ) In [33]: south Out[33]: 0 -0.494929 one 1.071804 2 0.721555 iii -0.706771 4 -i.039575 Proper noun: something, dtype: float64 In [34]: south . proper name Out[34]: 'something' The Series name will exist assigned automatically in many cases, in item when taking 1D slices of DataFrame as you volition see beneath.
Yous tin can rename a Serial with the pandas.Series.rename() method.
In [35]: s2 = due south . rename ( "different" ) In [36]: s2 . name Out[36]: 'unlike' Notation that s and s2 refer to different objects.
DataFrame¶
DataFrame is a 2-dimensional labeled data construction with columns of potentially different types. You can think of information technology like a spreadsheet or SQL table, or a dict of Series objects. It is by and large the nearly ordinarily used pandas object. Like Series, DataFrame accepts many different kinds of input:
-
Dict of 1D ndarrays, lists, dicts, or Series
-
2-D numpy.ndarray
-
Structured or record ndarray
-
A
Serial -
Another
DataFrame
Along with the data, you tin optionally pass index (row labels) and columns (column labels) arguments. If you pass an index and / or columns, you are guaranteeing the index and / or columns of the resulting DataFrame. Thus, a dict of Series plus a specific index will discard all data not matching up to the passed index.
If axis labels are non passed, they will exist constructed from the input data based on common sense rules.
Annotation
When the data is a dict, and columns is not specified, the DataFrame columns will exist ordered by the dict'due south insertion guild, if yous are using Python version >= three.6 and pandas >= 0.23.
If you are using Python < iii.half-dozen or pandas < 0.23, and columns is non specified, the DataFrame columns will be the lexically ordered list of dict keys.
From dict of Series or dicts¶
The resulting alphabetize will be the matrimony of the indexes of the various Series. If at that place are whatsoever nested dicts, these will kickoff be converted to Serial. If no columns are passed, the columns will be the ordered listing of dict keys.
In [37]: d = { ....: "1" : pd . Serial ([ 1.0 , 2.0 , 3.0 ], index = [ "a" , "b" , "c" ]), ....: "two" : pd . Serial ([ 1.0 , 2.0 , 3.0 , iv.0 ], index = [ "a" , "b" , "c" , "d" ]), ....: } ....: In [38]: df = pd . DataFrame ( d ) In [39]: df Out[39]: 1 2 a 1.0 ane.0 b 2.0 2.0 c three.0 3.0 d NaN 4.0 In [xl]: pd . DataFrame ( d , alphabetize = [ "d" , "b" , "a" ]) Out[40]: ane two d NaN four.0 b ii.0 two.0 a 1.0 1.0 In [41]: pd . DataFrame ( d , index = [ "d" , "b" , "a" ], columns = [ "two" , "3" ]) Out[41]: ii three d 4.0 NaN b 2.0 NaN a i.0 NaN The row and column labels tin can be accessed respectively by accessing the alphabetize and columns attributes:
Note
When a particular set up of columns is passed forth with a dict of information, the passed columns override the keys in the dict.
In [42]: df . index Out[42]: Alphabetize(['a', 'b', 'c', 'd'], dtype='object') In [43]: df . columns Out[43]: Index(['one', '2'], dtype='object') From dict of ndarrays / lists¶
The ndarrays must all exist the same length. If an alphabetize is passed, it must clearly likewise be the same length as the arrays. If no alphabetize is passed, the issue will be range(n) , where n is the array length.
In [44]: d = { "one" : [ 1.0 , 2.0 , 3.0 , 4.0 ], "2" : [ 4.0 , 3.0 , two.0 , one.0 ]} In [45]: pd . DataFrame ( d ) Out[45]: i two 0 1.0 4.0 1 two.0 iii.0 2 3.0 ii.0 3 4.0 1.0 In [46]: pd . DataFrame ( d , index = [ "a" , "b" , "c" , "d" ]) Out[46]: ane two a 1.0 4.0 b two.0 3.0 c 3.0 2.0 d four.0 1.0 From structured or tape array¶
This instance is handled identically to a dict of arrays.
In [47]: data = np . zeros (( two ,), dtype = [( "A" , "i4" ), ( "B" , "f4" ), ( "C" , "a10" )]) In [48]: data [:] = [( 1 , 2.0 , "Hello" ), ( ii , 3.0 , "World" )] In [49]: pd . DataFrame ( data ) Out[49]: A B C 0 one two.0 b'Hello' 1 2 3.0 b'Globe' In [l]: pd . DataFrame ( data , index = [ "commencement" , "2nd" ]) Out[l]: A B C first 1 2.0 b'Hello' 2nd two 3.0 b'Globe' In [51]: pd . DataFrame ( information , columns = [ "C" , "A" , "B" ]) Out[51]: C A B 0 b'Hello' 1 ii.0 1 b'World' 2 iii.0 Note
DataFrame is not intended to piece of work exactly like a 2-dimensional NumPy ndarray.
From a list of dicts¶
In [52]: data2 = [{ "a" : 1 , "b" : 2 }, { "a" : 5 , "b" : x , "c" : xx }] In [53]: pd . DataFrame ( data2 ) Out[53]: a b c 0 one ii NaN i 5 10 xx.0 In [54]: pd . DataFrame ( data2 , alphabetize = [ "first" , "second" ]) Out[54]: a b c first one two NaN second v ten 20.0 In [55]: pd . DataFrame ( data2 , columns = [ "a" , "b" ]) Out[55]: a b 0 ane 2 1 5 10 From a dict of tuples¶
Y'all can automatically create a MultiIndexed frame past passing a tuples lexicon.
In [56]: pd . DataFrame ( ....: { ....: ( "a" , "b" ): {( "A" , "B" ): ane , ( "A" , "C" ): two }, ....: ( "a" , "a" ): {( "A" , "C" ): 3 , ( "A" , "B" ): 4 }, ....: ( "a" , "c" ): {( "A" , "B" ): five , ( "A" , "C" ): 6 }, ....: ( "b" , "a" ): {( "A" , "C" ): vii , ( "A" , "B" ): viii }, ....: ( "b" , "b" ): {( "A" , "D" ): 9 , ( "A" , "B" ): ten }, ....: } ....: ) ....: Out[56]: a b b a c a b A B 1.0 4.0 v.0 8.0 10.0 C 2.0 3.0 6.0 7.0 NaN D NaN NaN NaN NaN ix.0 From a Series¶
The consequence volition be a DataFrame with the aforementioned index as the input Series, and with one column whose name is the original name of the Serial (merely if no other column proper name provided).
From a list of namedtuples¶
The field names of the first namedtuple in the list determine the columns of the DataFrame . The remaining namedtuples (or tuples) are merely unpacked and their values are fed into the rows of the DataFrame . If whatsoever of those tuples is shorter than the first namedtuple then the later columns in the corresponding row are marked as missing values. If any are longer than the showtime namedtuple , a ValueError is raised.
In [57]: from collections import namedtuple In [58]: Point = namedtuple ( "Point" , "ten y" ) In [59]: pd . DataFrame ([ Point ( 0 , 0 ), Signal ( 0 , three ), ( 2 , 3 )]) Out[59]: x y 0 0 0 ane 0 3 ii two three In [threescore]: Point3D = namedtuple ( "Point3D" , "x y z" ) In [61]: pd . DataFrame ([ Point3D ( 0 , 0 , 0 ), Point3D ( 0 , three , 5 ), Point ( 2 , iii )]) Out[61]: x y z 0 0 0 0.0 1 0 iii 5.0 ii 2 3 NaN From a list of dataclasses¶
New in version ane.1.0.
Data Classes as introduced in PEP557, can exist passed into the DataFrame constructor. Passing a list of dataclasses is equivalent to passing a list of dictionaries.
Please be enlightened, that all values in the list should be dataclasses, mixing types in the listing would result in a TypeError.
In [62]: from dataclasses import make_dataclass In [63]: Point = make_dataclass ( "Point" , [( "x" , int ), ( "y" , int )]) In [64]: pd . DataFrame ([ Betoken ( 0 , 0 ), Point ( 0 , 3 ), Betoken ( 2 , iii )]) Out[64]: x y 0 0 0 one 0 3 2 2 3 Missing data
Much more will exist said on this topic in the Missing data section. To construct a DataFrame with missing information, we use np.nan to represent missing values. Alternatively, you may pass a numpy.MaskedArray as the data argument to the DataFrame constructor, and its masked entries will exist considered missing.
Alternate constructors¶
DataFrame.from_dict
DataFrame.from_dict takes a dict of dicts or a dict of array-like sequences and returns a DataFrame. It operates like the DataFrame constructor except for the orient parameter which is 'columns' by default, merely which can be set to 'index' in order to use the dict keys as row labels.
In [65]: pd . DataFrame . from_dict ( dict ([( "A" , [ 1 , 2 , 3 ]), ( "B" , [ 4 , 5 , 6 ])])) Out[65]: A B 0 1 4 1 2 v two 3 6 If you pass orient='alphabetize' , the keys will be the row labels. In this case, you lot tin also pass the desired column names:
In [66]: pd . DataFrame . from_dict ( ....: dict ([( "A" , [ one , two , 3 ]), ( "B" , [ 4 , 5 , 6 ])]), ....: orient = "index" , ....: columns = [ "one" , "two" , "three" ], ....: ) ....: Out[66]: one 2 iii A 1 2 3 B 4 5 six DataFrame.from_records
DataFrame.from_records takes a list of tuples or an ndarray with structured dtype. Information technology works analogously to the normal DataFrame constructor, except that the resulting DataFrame index may be a specific field of the structured dtype. For example:
In [67]: data Out[67]: array([(1, 2., b'Howdy'), (2, 3., b'Globe')], dtype=[('A', '<i4'), ('B', '<f4'), ('C', 'S10')]) In [68]: pd . DataFrame . from_records ( data , index = "C" ) Out[68]: A B C b'Hello' 1 2.0 b'World' two 3.0 Cavalcade selection, improver, deletion¶
Yous can treat a DataFrame semantically like a dict of similar-indexed Serial objects. Getting, setting, and deleting columns works with the same syntax every bit the analogous dict operations:
In [69]: df [ "one" ] Out[69]: a one.0 b two.0 c three.0 d NaN Proper noun: i, dtype: float64 In [seventy]: df [ "3" ] = df [ "one" ] * df [ "two" ] In [71]: df [ "flag" ] = df [ "one" ] > ii In [72]: df Out[72]: ane two 3 flag a 1.0 1.0 i.0 Faux b 2.0 two.0 iv.0 False c 3.0 3.0 nine.0 True d NaN iv.0 NaN False Columns tin be deleted or popped similar with a dict:
In [73]: del df [ "ii" ] In [74]: 3 = df . pop ( "3" ) In [75]: df Out[75]: one flag a i.0 False b 2.0 False c 3.0 True d NaN False When inserting a scalar value, it will naturally be propagated to make full the column:
In [76]: df [ "foo" ] = "bar" In [77]: df Out[77]: one flag foo a one.0 False bar b 2.0 Faux bar c 3.0 True bar d NaN False bar When inserting a Series that does non accept the same index as the DataFrame, information technology will exist conformed to the DataFrame's index:
In [78]: df [ "one_trunc" ] = df [ "one" ][: 2 ] In [79]: df Out[79]: ane flag foo one_trunc a ane.0 Fake bar i.0 b 2.0 Fake bar 2.0 c 3.0 True bar NaN d NaN Fake bar NaN You can insert raw ndarrays but their length must match the length of the DataFrame's index.
By default, columns get inserted at the cease. The insert function is available to insert at a particular location in the columns:
In [eighty]: df . insert ( 1 , "bar" , df [ "one" ]) In [81]: df Out[81]: 1 bar flag foo one_trunc a 1.0 1.0 False bar 1.0 b 2.0 two.0 Faux bar 2.0 c iii.0 three.0 True bar NaN d NaN NaN Imitation bar NaN Assigning new columns in method chains¶
Inspired by dplyr's mutate verb, DataFrame has an assign() method that allows you to hands create new columns that are potentially derived from existing columns.
In [82]: iris = pd . read_csv ( "data/iris.information" ) In [83]: iris . head () Out[83]: SepalLength SepalWidth PetalLength PetalWidth Proper name 0 v.one three.5 ane.4 0.2 Iris-setosa 1 4.9 3.0 1.4 0.2 Iris-setosa ii 4.7 3.2 1.three 0.2 Iris-setosa 3 4.vi iii.1 1.v 0.2 Iris-setosa 4 5.0 iii.6 1.iv 0.2 Iris-setosa In [84]: iris . assign ( sepal_ratio = iris [ "SepalWidth" ] / iris [ "SepalLength" ]) . head () Out[84]: SepalLength SepalWidth PetalLength PetalWidth Proper name sepal_ratio 0 5.one iii.5 1.4 0.two Iris-setosa 0.686275 1 4.nine iii.0 1.4 0.ii Iris-setosa 0.612245 2 four.seven 3.2 1.3 0.2 Iris-setosa 0.680851 3 4.6 3.1 ane.5 0.2 Iris-setosa 0.673913 4 5.0 3.half-dozen one.4 0.ii Iris-setosa 0.720000 In the example in a higher place, nosotros inserted a precomputed value. Nosotros can also pass in a function of one argument to be evaluated on the DataFrame being assigned to.
In [85]: iris . assign ( sepal_ratio = lambda 10 : ( x [ "SepalWidth" ] / x [ "SepalLength" ])) . caput () Out[85]: SepalLength SepalWidth PetalLength PetalWidth Proper name sepal_ratio 0 5.1 three.5 one.4 0.ii Iris-setosa 0.686275 1 iv.9 3.0 1.4 0.ii Iris-setosa 0.612245 2 four.vii iii.2 i.iii 0.2 Iris-setosa 0.680851 3 4.vi 3.ane ane.5 0.2 Iris-setosa 0.673913 4 5.0 3.vi one.four 0.2 Iris-setosa 0.720000 assign always returns a re-create of the information, leaving the original DataFrame untouched.
Passing a callable, as opposed to an actual value to be inserted, is useful when you don't take a reference to the DataFrame at hand. This is mutual when using assign in a chain of operations. For case, we can limit the DataFrame to just those observations with a Sepal Length greater than 5, calculate the ratio, and plot:
In [86]: ( ....: iris . query ( "SepalLength > five" ) ....: . assign ( ....: SepalRatio = lambda 10 : ten . SepalWidth / x . SepalLength , ....: PetalRatio = lambda x : x . PetalWidth / x . PetalLength , ....: ) ....: . plot ( kind = "scatter" , 10 = "SepalRatio" , y = "PetalRatio" ) ....: ) ....: Out[86]: <AxesSubplot:xlabel='SepalRatio', ylabel='PetalRatio'> 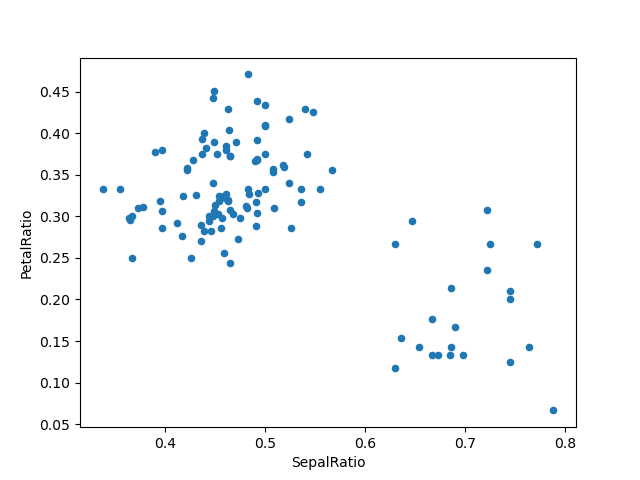
Since a function is passed in, the function is computed on the DataFrame beingness assigned to. Chiefly, this is the DataFrame that's been filtered to those rows with sepal length greater than 5. The filtering happens starting time, and so the ratio calculations. This is an example where we didn't have a reference to the filtered DataFrame available.
The function signature for assign is just **kwargs . The keys are the column names for the new fields, and the values are either a value to be inserted (for example, a Series or NumPy array), or a function of ane argument to exist called on the DataFrame . A copy of the original DataFrame is returned, with the new values inserted.
Starting with Python 3.6 the order of **kwargs is preserved. This allows for dependent consignment, where an expression later in **kwargs can refer to a column created earlier in the same assign() .
In [87]: dfa = pd . DataFrame ({ "A" : [ ane , 2 , three ], "B" : [ 4 , v , 6 ]}) In [88]: dfa . assign ( C = lambda 10 : x [ "A" ] + x [ "B" ], D = lambda x : ten [ "A" ] + 10 [ "C" ]) Out[88]: A B C D 0 1 four v 6 1 ii 5 7 ix 2 iii 6 9 12 In the second expression, x['C'] will refer to the newly created cavalcade, that's equal to dfa['A'] + dfa['B'] .
Indexing / selection¶
The basics of indexing are equally follows:
| Performance | Syntax | Effect |
|---|---|---|
| Select cavalcade | | Serial |
| Select row by label | | Series |
| Select row by integer location | | Serial |
| Piece rows | | DataFrame |
| Select rows by boolean vector | | DataFrame |
Row selection, for example, returns a Series whose index is the columns of the DataFrame:
In [89]: df . loc [ "b" ] Out[89]: 1 two.0 bar 2.0 flag Simulated foo bar one_trunc ii.0 Name: b, dtype: object In [90]: df . iloc [ 2 ] Out[90]: i iii.0 bar 3.0 flag True foo bar one_trunc NaN Name: c, dtype: object For a more than exhaustive treatment of sophisticated label-based indexing and slicing, see the section on indexing. Nosotros will address the fundamentals of reindexing / befitting to new sets of labels in the section on reindexing.
Data alignment and arithmetic¶
Information alignment between DataFrame objects automatically align on both the columns and the index (row labels). Again, the resulting object will have the matrimony of the cavalcade and row labels.
In [91]: df = pd . DataFrame ( np . random . randn ( 10 , 4 ), columns = [ "A" , "B" , "C" , "D" ]) In [92]: df2 = pd . DataFrame ( np . random . randn ( 7 , 3 ), columns = [ "A" , "B" , "C" ]) In [93]: df + df2 Out[93]: A B C D 0 0.045691 -0.014138 1.380871 NaN i -0.955398 -1.501007 0.037181 NaN two -0.662690 1.534833 -0.859691 NaN 3 -2.452949 1.237274 -0.133712 NaN four 1.414490 1.951676 -2.320422 NaN 5 -0.494922 -i.649727 -1.084601 NaN 6 -i.047551 -0.748572 -0.805479 NaN seven NaN NaN NaN NaN 8 NaN NaN NaN NaN nine NaN NaN NaN NaN When doing an operation between DataFrame and Series, the default beliefs is to align the Series index on the DataFrame columns, thus broadcasting row-wise. For example:
In [94]: df - df . iloc [ 0 ] Out[94]: A B C D 0 0.000000 0.000000 0.000000 0.000000 1 -1.359261 -0.248717 -0.453372 -1.754659 2 0.253128 0.829678 0.010026 -i.991234 3 -one.311128 0.054325 -i.724913 -1.620544 iv 0.573025 1.500742 -0.676070 1.367331 5 -ane.741248 0.781993 -one.241620 -two.053136 six -i.240774 -0.869551 -0.153282 0.000430 vii -0.743894 0.411013 -0.929563 -0.282386 8 -one.194921 1.320690 0.238224 -1.482644 9 2.293786 ane.856228 0.773289 -ane.446531 For explicit control over the matching and broadcasting behavior, see the section on flexible binary operations.
Operations with scalars are only as yous would await:
In [95]: df * v + two Out[95]: A B C D 0 three.359299 -0.124862 iv.835102 three.381160 ane -iii.437003 -1.368449 2.568242 -v.392133 2 4.624938 4.023526 4.885230 -6.575010 iii -3.196342 0.146766 -3.789461 -iv.721559 4 6.224426 7.378849 1.454750 10.217815 5 -5.346940 3.785103 -1.373001 -6.884519 6 -2.844569 -four.472618 four.068691 three.383309 7 -0.360173 1.930201 0.187285 i.969232 8 -2.615303 6.478587 6.026220 -4.032059 9 14.828230 nine.156280 8.701544 -three.851494 In [96]: ane / df Out[96]: A B C D 0 3.678365 -2.353094 1.763605 iii.620145 ane -0.919624 -1.484363 8.799067 -0.676395 ii one.904807 2.470934 1.732964 -0.583090 three -0.962215 -2.697986 -0.863638 -0.743875 4 1.183593 0.929567 -9.170108 0.608434 five -0.680555 2.800959 -one.482360 -0.562777 6 -1.032084 -0.772485 2.416988 3.614523 7 -ii.118489 -71.634509 -two.758294 -162.507295 8 -1.083352 1.116424 one.241860 -0.828904 nine 0.389765 0.698687 0.746097 -0.854483 In [97]: df ** four Out[97]: A B C D 0 0.005462 three.261689e-02 0.103370 5.822320e-03 1 ane.398165 2.059869e-01 0.000167 4.777482e+00 2 0.075962 two.682596e-02 0.110877 8.650845e+00 3 1.166571 one.887302e-02 1.797515 3.265879e+00 4 0.509555 one.339298e+00 0.000141 vii.297019e+00 5 iv.661717 1.624699e-02 0.207103 9.969092e+00 6 0.881334 two.808277e+00 0.029302 5.858632e-03 7 0.049647 3.797614e-08 0.017276 1.433866e-09 8 0.725974 6.437005e-01 0.420446 ii.118275e+00 ix 43.329821 iv.196326e+00 3.227153 1.875802e+00 Boolean operators work every bit well:
In [98]: df1 = pd . DataFrame ({ "a" : [ ane , 0 , 1 ], "b" : [ 0 , 1 , 1 ]}, dtype = bool ) In [99]: df2 = pd . DataFrame ({ "a" : [ 0 , 1 , 1 ], "b" : [ 1 , 1 , 0 ]}, dtype = bool ) In [100]: df1 & df2 Out[100]: a b 0 Faux False one Fake Truthful 2 True False In [101]: df1 | df2 Out[101]: a b 0 True True i True Truthful ii Truthful Truthful In [102]: df1 ^ df2 Out[102]: a b 0 True True 1 True False 2 False Truthful In [103]: - df1 Out[103]: a b 0 Fake True ane True False 2 False False Transposing¶
To transpose, access the T attribute (besides the transpose function), like to an ndarray:
# only show the get-go five rows In [104]: df [: v ] . T Out[104]: 0 1 2 3 4 A 0.271860 -1.087401 0.524988 -1.039268 0.844885 B -0.424972 -0.673690 0.404705 -0.370647 1.075770 C 0.567020 0.113648 0.577046 -1.157892 -0.109050 D 0.276232 -1.478427 -ane.715002 -1.344312 1.643563 DataFrame interoperability with NumPy functions¶
Elementwise NumPy ufuncs (log, exp, sqrt, …) and various other NumPy functions can be used with no problems on Series and DataFrame, assuming the data inside are numeric:
In [105]: np . exp ( df ) Out[105]: A B C D 0 1.312403 0.653788 1.763006 ane.318154 1 0.337092 0.509824 one.120358 0.227996 2 1.690438 1.498861 1.780770 0.179963 3 0.353713 0.690288 0.314148 0.260719 4 2.327710 2.932249 0.896686 5.173571 5 0.230066 1.429065 0.509360 0.169161 6 0.379495 0.274028 one.512461 1.318720 7 0.623732 0.986137 0.695904 0.993865 8 0.397301 2.449092 two.237242 0.299269 9 xiii.009059 iv.183951 3.820223 0.310274 In [106]: np . asarray ( df ) Out[106]: array([[ 0.2719, -0.425 , 0.567 , 0.2762], [-1.0874, -0.6737, 0.1136, -1.4784], [ 0.525 , 0.4047, 0.577 , -1.715 ], [-1.0393, -0.3706, -1.1579, -1.3443], [ 0.8449, one.0758, -0.109 , one.6436], [-1.4694, 0.357 , -0.6746, -1.7769], [-0.9689, -1.2945, 0.4137, 0.2767], [-0.472 , -0.014 , -0.3625, -0.0062], [-0.9231, 0.8957, 0.8052, -1.2064], [ two.5656, 1.4313, 1.3403, -ane.1703]]) DataFrame is not intended to be a drop-in replacement for ndarray equally its indexing semantics and data model are quite different in places from an n-dimensional array.
Series implements __array_ufunc__ , which allows information technology to work with NumPy's universal functions.
The ufunc is applied to the underlying assortment in a Series.
In [107]: ser = pd . Series ([ ane , two , 3 , 4 ]) In [108]: np . exp ( ser ) Out[108]: 0 ii.718282 1 7.389056 2 twenty.085537 3 54.598150 dtype: float64 Changed in version 0.25.0: When multiple Series are passed to a ufunc, they are aligned before performing the operation.
Like other parts of the library, pandas will automatically align labeled inputs as function of a ufunc with multiple inputs. For example, using numpy.balance() on ii Series with differently ordered labels will align before the operation.
In [109]: ser1 = pd . Series ([ ane , ii , 3 ], index = [ "a" , "b" , "c" ]) In [110]: ser2 = pd . Serial ([ 1 , 3 , 5 ], index = [ "b" , "a" , "c" ]) In [111]: ser1 Out[111]: a one b 2 c 3 dtype: int64 In [112]: ser2 Out[112]: b 1 a 3 c 5 dtype: int64 In [113]: np . remainder ( ser1 , ser2 ) Out[113]: a ane b 0 c 3 dtype: int64 As usual, the matrimony of the two indices is taken, and not-overlapping values are filled with missing values.
In [114]: ser3 = pd . Serial ([ 2 , iv , 6 ], index = [ "b" , "c" , "d" ]) In [115]: ser3 Out[115]: b 2 c iv d vi dtype: int64 In [116]: np . residual ( ser1 , ser3 ) Out[116]: a NaN b 0.0 c 3.0 d NaN dtype: float64 When a binary ufunc is applied to a Series and Index , the Series implementation takes precedence and a Series is returned.
In [117]: ser = pd . Series ([ 1 , 2 , three ]) In [118]: idx = pd . Index ([ iv , 5 , 6 ]) In [119]: np . maximum ( ser , idx ) Out[119]: 0 4 one v 2 6 dtype: int64 NumPy ufuncs are safe to apply to Series backed by non-ndarray arrays, for example arrays.SparseArray (come across Thin adding). If possible, the ufunc is practical without converting the underlying data to an ndarray.
Console display¶
Very large DataFrames will be truncated to brandish them in the panel. Yous tin can besides get a summary using info() . (Here I am reading a CSV version of the baseball dataset from the plyr R package):
In [120]: baseball game = pd . read_csv ( "data/baseball game.csv" ) In [121]: print ( baseball game ) id role player yr stint team lg m ab r h X2b X3b 60 minutes rbi sb cs bb so ibb hbp sh sf gidp 0 88641 womacto01 2006 ii CHN NL 19 fifty half-dozen 14 1 0 1 2.0 1.0 one.0 4 four.0 0.0 0.0 3.0 0.0 0.0 1 88643 schilcu01 2006 i BOS AL 31 2 0 i 0 0 0 0.0 0.0 0.0 0 i.0 0.0 0.0 0.0 0.0 0.0 .. ... ... ... ... ... .. .. ... .. ... ... ... .. ... ... ... .. ... ... ... ... ... ... 98 89533 aloumo01 2007 1 NYN NL 87 328 51 112 19 i 13 49.0 3.0 0.0 27 30.0 v.0 2.0 0.0 3.0 13.0 99 89534 alomasa02 2007 one NYN NL eight 22 1 3 1 0 0 0.0 0.0 0.0 0 three.0 0.0 0.0 0.0 0.0 0.0 [100 rows x 23 columns] In [122]: baseball . info () <grade 'pandas.core.frame.DataFrame'> RangeIndex: 100 entries, 0 to 99 Data columns (total 23 columns): # Column Not-Null Count Dtype --- ------ -------------- ----- 0 id 100 not-null int64 1 player 100 non-zilch object ii year 100 non-null int64 iii stint 100 non-cipher int64 4 team 100 not-null object 5 lg 100 non-cypher object 6 g 100 not-null int64 vii ab 100 non-null int64 8 r 100 non-nix int64 9 h 100 non-goose egg int64 ten X2b 100 non-null int64 11 X3b 100 non-null int64 12 hr 100 non-naught int64 xiii rbi 100 not-cipher float64 xiv sb 100 non-zippo float64 15 cs 100 not-null float64 sixteen bb 100 non-nada int64 17 so 100 not-cipher float64 18 ibb 100 non-null float64 19 hbp 100 non-zippo float64 twenty sh 100 not-null float64 21 sf 100 non-null float64 22 gidp 100 non-zilch float64 dtypes: float64(9), int64(11), object(three) retention usage: 18.1+ KB However, using to_string will return a string representation of the DataFrame in tabular grade, though it won't always fit the panel width:
In [123]: impress ( baseball . iloc [ - xx :, : 12 ] . to_string ()) id player year stint team lg g ab r h X2b X3b 80 89474 finlest01 2007 i COL NL 43 94 9 17 iii 0 81 89480 embreal01 2007 1 OAK AL iv 0 0 0 0 0 82 89481 edmonji01 2007 ane SLN NL 117 365 39 92 15 2 83 89482 easleda01 2007 1 NYN NL 76 193 24 54 6 0 84 89489 delgaca01 2007 1 NYN NL 139 538 71 139 xxx 0 85 89493 cormirh01 2007 ane CIN NL 6 0 0 0 0 0 86 89494 coninje01 2007 two NYN NL 21 41 2 8 2 0 87 89495 coninje01 2007 1 CIN NL 80 215 23 57 11 i 88 89497 clemero02 2007 1 NYA AL two 2 0 i 0 0 89 89498 claytro01 2007 2 BOS AL 8 6 1 0 0 0 90 89499 claytro01 2007 1 TOR AL 69 189 23 48 fourteen 0 91 89501 cirilje01 2007 2 ARI NL 28 40 6 viii 4 0 92 89502 cirilje01 2007 1 MIN AL l 153 18 forty 9 two 93 89521 bondsba01 2007 1 SFN NL 126 340 75 94 14 0 94 89523 biggicr01 2007 1 HOU NL 141 517 68 130 31 3 95 89525 benitar01 2007 2 FLO NL 34 0 0 0 0 0 96 89526 benitar01 2007 one SFN NL nineteen 0 0 0 0 0 97 89530 ausmubr01 2007 1 HOU NL 117 349 38 82 xvi iii 98 89533 aloumo01 2007 1 NYN NL 87 328 51 112 nineteen 1 99 89534 alomasa02 2007 1 NYN NL 8 22 1 3 ane 0 Broad DataFrames will be printed across multiple rows past default:
In [124]: pd . DataFrame ( np . random . randn ( three , 12 )) Out[124]: 0 ane 2 3 4 5 6 7 8 9 10 eleven 0 -one.226825 0.769804 -1.281247 -0.727707 -0.121306 -0.097883 0.695775 0.341734 0.959726 -1.110336 -0.619976 0.149748 1 -0.732339 0.687738 0.176444 0.403310 -0.154951 0.301624 -2.179861 -1.369849 -0.954208 1.462696 -i.743161 -0.826591 2 -0.345352 1.314232 0.690579 0.995761 2.396780 0.014871 3.357427 -0.317441 -1.236269 0.896171 -0.487602 -0.082240 Y'all can change how much to print on a unmarried row by setting the display.width option:
In [125]: pd . set_option ( "display.width" , 40 ) # default is lxxx In [126]: pd . DataFrame ( np . random . randn ( three , 12 )) Out[126]: 0 1 2 3 4 5 half dozen 7 viii 9 x 11 0 -two.182937 0.380396 0.084844 0.432390 1.519970 -0.493662 0.600178 0.274230 0.132885 -0.023688 two.410179 1.450520 1 0.206053 -0.251905 -two.213588 i.063327 1.266143 0.299368 -0.863838 0.408204 -1.048089 -0.025747 -0.988387 0.094055 2 1.262731 i.289997 0.082423 -0.055758 0.536580 -0.489682 0.369374 -0.034571 -2.484478 -0.281461 0.030711 0.109121 Yous can adapt the max width of the individual columns by setting display.max_colwidth
In [127]: datafile = { .....: "filename" : [ "filename_01" , "filename_02" ], .....: "path" : [ .....: "media/user_name/storage/folder_01/filename_01" , .....: "media/user_name/storage/folder_02/filename_02" , .....: ], .....: } .....: In [128]: pd . set_option ( "brandish.max_colwidth" , 30 ) In [129]: pd . DataFrame ( datafile ) Out[129]: filename path 0 filename_01 media/user_name/storage/fo... ane filename_02 media/user_name/storage/fo... In [130]: pd . set_option ( "display.max_colwidth" , 100 ) In [131]: pd . DataFrame ( datafile ) Out[131]: filename path 0 filename_01 media/user_name/storage/folder_01/filename_01 one filename_02 media/user_name/storage/folder_02/filename_02 Y'all can likewise disable this feature via the expand_frame_repr option. This will print the table in 1 block.
DataFrame column aspect admission and IPython completion¶
If a DataFrame column label is a valid Python variable proper noun, the column can be accessed like an attribute:
In [132]: df = pd . DataFrame ({ "foo1" : np . random . randn ( five ), "foo2" : np . random . randn ( 5 )}) In [133]: df Out[133]: foo1 foo2 0 ane.126203 0.781836 ane -0.977349 -1.071357 ii 1.474071 0.441153 3 -0.064034 2.353925 4 -ane.282782 0.583787 In [134]: df . foo1 Out[134]: 0 i.126203 1 -0.977349 ii 1.474071 3 -0.064034 4 -one.282782 Name: foo1, dtype: float64 The columns are also connected to the IPython completion mechanism so they tin be tab-completed:
In [5]: df . foo < TAB > # noqa: E225, E999 df.foo1 df.foo2 How to Read Wikipedia Data to Pandas Dataframe
Source: https://pandas.pydata.org/pandas-docs/stable/user_guide/dsintro.html